Furfural is a heterocyclic aldehyde derived from furan. It is an oily, pale yellow liquid [1] with a tendency to darkening - photochemical oxidation - in the presence of light and oxygen [2]. It is a recalcitrant and toxic substance to various living organisms and severely inhibits the enzymatic metabolism of many microorganisms. In any case, furfural can be eliminated by using the capacity of some bacteria to metabolize it [3].
Widely used by chemical industries, furfural acts as an intermediate product for the manufacture of plastics, paint solvents, furan resin and adhesives, among other applications. However, despite its production and exportation benefits, it is known to cause serious environmental problems during its production process [4].
Many technologies are now available to treat effluents that have been contaminated with hazardous materials such as furfural. Among these options, biodegradation stands out, gaining increasing attention owing to its efficient biotechnological approach to restore contaminated environments [5]. It involves the use of microorganisms to degrade environmental pollutants. In this regard, some studies have engineered furfural metabolizing strains for the biological detoxification of the furan aldehydes by cloning or adapting the furfural metabolic pathway from bacteria such as C. basilensis [3], P. putida [6] and E. coli strains [7].
The genus Microbacterium is part of the Phylum Actinobacteria, a group of Gram-positive bacteria present in high concentrations in soils and aquatic sediments. Not only can they grow and subsist in very diverse substrates but also, they can metabolize complex organic matter [8], playing an important role in the recycling of substances. It has also been demonstrated that Actinobacteria play an important ecological role thanks to their ability to eliminate xenobiotic compounds such as pesticides [9] and heavy metals [10], among other substances. Khan et al. [11] determined that the group of Actinobacteria was one of the most abundant microorganisms in the bacterial community structure in anaerobic digesters of a full-scale municipal wastewater treatment plant. Haleyur et al. [12] used Microbacterium sp. in a consortium as a successful Polycyclic Aromatic Hydrocarbons (PAHs) degrader.
By contrast, Bacillus genus microorganisms are large-sized bacilli (4 – 10 μm), Gram-positive, strict aerobic or encapsulated facultative anaerobes. Bacteria are ubiquitous in soils, some of them are species of economic value due to their production of compounds such as enzymes, antibiotics and surfactants used for various industrial purposes. The ability of Bacillus species of forming highly resistant endospores gives them a very important competitive advantage in an environment such as soil. In addition to that, they can adapt to sudden temperature changes because of an inducible heat shock gene that includes chaperone proteins and proteases [13]. The Bacillus spores are generally formed under limited nutrients conditions [14]. Different studies demonstrated the ability of some Bacillus species to form bacterial consortiums able to degrade oil [15], lubricating oils residues [16] and petroleum hydrocarbons in crude oil contaminated soil [17]. Felshia et al. [18] identified strains of Bacillus licheniformis which demonstrate phenol degradation up to 2000 mg L-1. Yan et al. [19], found a bio-catalytic reduction of furfural to furfuryl alcohol by using Bacillus coagulans, a furfural tolerant strain, and glucose as co-substrate.
Brevundimonas are a genus of proteobacteria, Gram-negative, non-fermentative aerobic bacilli. Brevundimonas species are ubiquitous in the environment and some clones have been recovered from hydrocarbons contaminated soils [20]. The genus is known for the degradation of a variety of phenolic contaminants [21] and some of its species can make use of diesel oil as the sole source of carbon and energy [22]. Some species have been associated with a potential and activity of polyaromatic hydrocarbons degradation increase [23]. Basuki [24] identified strains of Brevundimonasdiminuta capable of significantly degrading aliphatic, aromatic and polycyclic hydrocarbons of short and long chains.
In previous studies, sludge bacteria were isolated from the tannin effluent treatment plant and 500 mg L-1 of furfural was used as a selection pressure [25]. The unidentified consortium was able to grow and degrade furfural up to 600 mg L-1 in 20 hours [26].
The microorganisms identified by the Polymerase Chain Reaction amplification method (PCR) and sequencing of the 16S rRNA (Ribosomal Nucleic Acid) gene were, among others, Microbacterium sp., Bacillus licheniformis and Brevundimonas sp. Degradation of furfural by the bacteria was determined individually, achieving a removal of up to 1300 mg L-1 of this compound
The aim of this paper was to determine the ability of these bacteria to act in consortium and remove furfural at high contaminant concentrations.
Strains of Microbacterium sp., Bacillus licheniformis and Brevundimonas sp., previously isolated, identified by PCR-amplification and 16S-rRNA-gene-sequencing method and properly conserved, were used for furfural degradation tests.
The growth of bacterial strains was carried out in two stages: meat peptone solution inoculation and subsequent minimal medium (M9) inoculation. The solutions and materials used were previously autoclaved at 1.5 bar and 112 °C for 15 minutes.
The first inoculum was made in three 250 mL Erlenmeyer flasks (one for each strain), containing 49 mL of meat peptone to a 1% m/v, 0.5 mL of NH4Cl 1 M (JT Baker Inc., 99.7% purity), 0.5 mL of NaH2PO4 1 M (Mallinckrodt Chemical Works of 99.0% purity) each. Colonies were sown from a solid medium (nutritive agar) with 24 hours of development (Figure 1).
These enrichments were incubated at 30 ºC using a DLAB orbital shaker (SK-0330-PRO) at 200 rpm, in darkness for 48 h.
Subsequently, a second inoculum was carried out in a minimal mineral medium consisting of 32 mL of sterile deionized water, 10 mL of M9, prepared with 34 g of sodium monoacid phosphate (Na2HPO4) (Mallinckrodt Chemical Works, 99.8% purity), 15 g of potassium diacid phosphate (KH2PO4) (Mallinckrodt Chemical Works, 99.0% purity), 2.5 g of sodium chloride (NaCl) (Alcor, 99.9% purity) and 0.5 g of ammonium chloride ((NH4Cl) (JT Baker Inc., 99.7% pure), adjusted to 1 liter. It was then planted with 2.5 mL of glucose (C6H12O6) (Cicarelli, 99.0%) at 20% m/v, 0.1 mL of 1M magnesium sulfate (MgSO4) (Cicarelli, 98.0% purity) and 5 µL of calcium chloride (CaCl2) 1M (Merck, 99.5% pure).
5 mL of meat peptone inoculum was added and incubated at 30 °C on a DLAB orbital shaker (SK-0330-PRO) at 200 rpm, for 24 hours.
Left: Bacillus licheniformis; center: Microbacterium sp.; right: Brevundimonas sp.
A triplicated culture was made. Erlenmeyer flasks of 250 mL were prepared with: 34 mL of sterile distilled water, 10 mL of M9, 0.1 mL of magnesium sulfate 1 M (Cicarelli, 98.0% purity), 5 µL of calcium chloride 1 M, 1 mL of glucose at 20% m/v.
The consortium was inoculated as follows: 1.7 mL of Microbacterium sp. inoculum, 1.7 mL of the inoculum of Bacillus licheniformis and 1.7 mL of the inoculum of Brevundimonas sp.; each one of them at 0.7 optical density at 610 nm (OD610), thus estimating a total amount of bacteria of 106 Colony Forming Units (CFU). Eight furfural concentrations between 722 and 4517 mg L-1 were tested.
An abiotic control was performed at the same time under the same cultivation conditions.
The flasks were incubated at 30 °C at 200 rpm for 72 h using a DLAB orbital shaker (SK-0330-PRO). Bacterial growth was monitored by measuring culture turbidity at 610 nm with a PerkinElmer UV / VIS Lambda 25 spectrophotometer.
Furfural concentration was analyzed every 24 hours by High Performance Liquid Chromatography (HPLC) using a Shimadzu CBM 20A device with UV SPD 20A detector at 278 nm wavelength, and with a C18 reverse phase column, at 40 °C, with 1 mL min-1 flow rate. In addition, an injection volume of 20 μL acetonitrile – water (17.5: 82.5) mixture was used as mobile phase [27].
In order to determine the furfural inhibitory concentration for the consortium, 722, 964, 1365, 1488, 1749, 1990, 2724, 4517 mg L-1 were tested. Furfural consumption was detected in all trials. Figure 2 shows that bacterial consortium formed by the strains of Microbacterium sp., Brevundimonas sp. and Bacillus licheniformis is capable of metabolizing the contaminant in 72 hours, at least up to 4517 mg L-1 yet, in the latter case, with lower furfural consumption. From 1749 mg L-1 to 2724 mg L-1, total contaminant degradation was not achieved in 24 hours. Conversely, furfural was completely degraded in 48 hours in this range of concentration. A reduction in consumption was observed at high concentration of pollutant (greater than 4500 mg L-1), but full degradation was not observed in three days.
Farías et al. [28] isolated the Bacillus licheniformis strain and tested it with furfural concentrations of 1400 mg L-1. As a result, it was demonstrated that this strain tolerated this condition and degraded the pollutant by 11.8% in 3 days.
Instead, in the study by Zheng et al. [2], Bacillus cereus sp. showed that at a furfural concentration of 500 mg L-1, the best furfural degradation ability was 35% in 7 days.
Furfural degradation by bacterial consortium vs Time, incubation conditions 30 º C at 200 rpm; c0 indicates initial furfural concentration
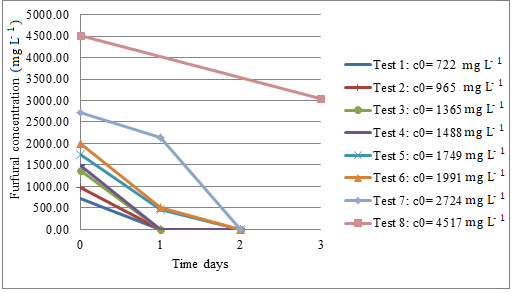
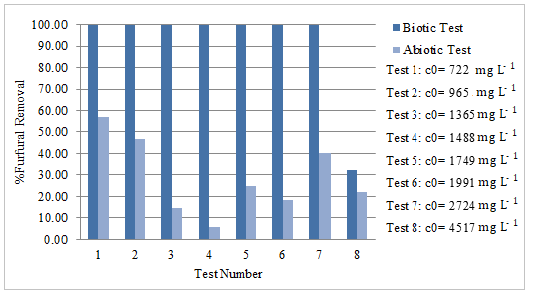
Figure 3 shows furfural´s removal percentage at 72 hours, related to the abiotic test. The average abiotic degradation was 28%. In all cases, a significantly higher degradation is observed in biotic essays. The degradation of 100% of the pollutant was achieved up to concentrations of 2723 mg L-1 and 11% effective (biotic least abiotic) at concentrations of 4516 mg L-1. It was observed, nonetheless, that it was difficult for the consortium to grow and the furfural to be metabolized at higher concentrations. 11 % of the contaminant was removed in 72 hours, which suggests that it may be possible to achieve the complete degradation in longer periods.
Boopathy [5] found a growth limit for Methanococcus sp. at 2400 mg L-1 of furfural concentration and 100% degradation at lower concentrations but in a 5 days incubation period. Zheng et al. [29] isolated Bacillus subtilis strain DS3, which is capable of growing on furfural as sole carbon source. Despite the similarities in the results, degradation percentages were lower (about 30%) in longer periods (7 days).
Degradation of furfural in abiotic tests can be observed, which may suggest that this decrease could be a photochemical degradation of furfural as mentioned by Zheng et al. [2] and not a result from bacterial action. Similarly, Bariani et al. [30] and Steinbach et al. [31], report furfural degradation reactions that lead to autoxidation when it is exposed to air. Nevertheless, bacterial degradation is clearly observable.
The bacterial growth was then monitored from the absorbance measured at 610 nm.
In previous studies (data not shown), the capacity of both Brevundimonas sp. and Microbacterium sp. to grow in a furfural environment was determined. About 1400 mg L-1, OD610 readings showed less than 0.15 after 3 days. It can be inferred that these microorganisms have not been able to metabolize furfural in that time.
Furfural removal percentage at 3 days in biotic and abiotic test, incubation conditions 30 º C at 200 rpm; c0 indicates initial furfural concentration
The capacity of each microorganism alone to grow in a furfural environment and use it as the sole carbon and energy source was determined by some authors.
Table 1 shows the impact of furfural concentration in individual bacterial growth during days. It displays a complete inhibition in individual strains with furfural initial concentration at 1433 mg L-1, specifically on Bacillus licheniformis strain. Both, L. mesenteroidesand L. pseudomesenteroideshave achieved growth at 2200 mg L-1 of furfural with OD610 levels between 0.46 and 0.62 in 3 days. Similar results were obtained with Z. mobilis at 1900 mg L-1. As regards Bacillus cereus sp. and Methanococcus sp., they only achieved 0.30 OD610 in 7 days. Our consortium could grow at 2724 mg L-1 of furfural. It can be seen in the high OD610 level (1.81) in only 3 days.
Furfural growth inhibition; OD values of individual strains vs consortium
|
Bacteria |
Furfural initial concentr. (mg L-1) |
Absorbances |
||||
|
day 0 |
day 1 |
day 2 |
day 3 |
day 7 |
||
|
B. licheniformis (Farías et al., 2019) |
1400 |
0.11 |
0.10 |
0.10 |
0.10 |
- |
|
Microbacterium sp. (Echeverría et al., 2018) |
1600 |
0.10 |
0.10 |
0.11 |
0.14 |
- |
|
Z. mobilis (Hunter and Manter, 2014) |
1900 |
0.05 |
0.40 |
0.60 |
0.60 |
- |
|
L. mesenteroides (ibid.) |
2200 |
0.09 |
0.45 |
0.55 |
0.62 |
- |
|
L. pseudomesenteroides (ibid.) |
2200 |
0.09 |
0.30 |
0.42 |
0.46 |
- |
|
Bacillus cereus sp. (Zheng et al., 2015) |
2000 |
- |
- |
- |
- |
0.32 |
|
Methanococcus sp.(Boopathy et al., 1993) |
2000 |
0.02 |
0.03 |
0.10 |
0.16 |
0.27 |
|
Consortium |
2700 |
0.14 |
0.23 |
1.81 |
- |
- |
Several studies showed significant differences between microbial consortium and individual strains concerning advantage in their degradative capacities or in various biotechnological processes. Thus, Ding et al. [32] indicated the degradation effects for Chemical Oxygen Demand determined by the Dichromate Method (CODCr) of single isolated strain, mixed strains, and microbial consortium originally screened from viscose fiber wastewater. Microbial consortium showed the optimum degradation rate of CODCr. Jing et al. [33] made a comparison between individual strains and the microbial consortium for biopolymer production in different treatment processes. The experimental results showed that the microbial consortium could produce more biopolymers than individual strains, and the reason might be attributed to the synergetic action of strains. Jing et al. [34] compared the degradation of phenanthrene between individual predominant strains and the microbial consortium in different treatment processes. The microbial consortium showed a significant improvement of phenanthrene degradation rates in either static or shaken culture.
Boopathy [5] indicated that furfural is a germicide. This means that it inhibits the growth of some biological systems. This effect has also been reported by Moussavi et al. [35] who made a degradation test in a continuous inflow cyclic biological reactor (CBR) and could remove over 98% of furfural at inlet furfural concentration up to 1200 mg L-1. The studies carried out by Crigler et al. [7] characterized the genes involved in the aerobic bacterial metabolism of furfural in two species, Pseudomonas putida Fu1 and Cupriavidusbasilensis HMF14.
Even though at higher concentrations, a difficulty in the consortium growing and furfural metabolizing was observed, the removal of 11% of the contaminant in 72 hours, previously described, suggests the possibility of its degradation in longer periods. Hunter and Manter [36] showed that the furfural treatments slowed the growth rates of three L. mesenteroides and L. pseudomesenteroidesstrains, but they obtained better degradation values (between 55% to 75% at 4500 mg L-1 of furfural).
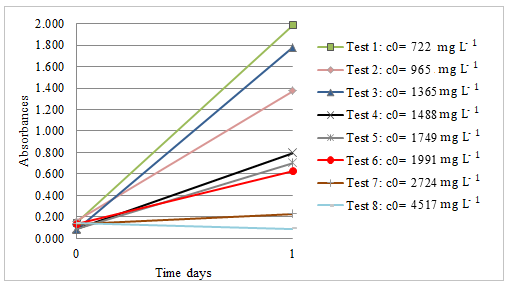
Figure 4 represents 24 h bacterial growth, according to different furfural concentrations. The bacterial consortium growth was monitored by measuring culture turbidity at 610 nm. Up to 1991 mg L-1, the bacterial consortium was able to grow rapidly in just 24 hours (OD610 ˃ 0.6). However, the influence of furfural concentration on bacterial growth can be seen due to the turbidity decrease in solutions with increasing pollutant concentration. Consequently, decreasing slopes at 4517 mg L-1 were observed. The results could give an idea of the amount of viable cells needed to degrade furfural at those particular concentrations.
Although, Guarnieri et al. [37] obtained through genetic engineering, a P. putida KT2440 strain capable of growth on furfural as a sole carbon and energy source up to 1000 mg L-1, that bacteria needed more than 15 hours to reach 0.6 OD610, while our consortium could grow in similar concentrations of furfural, achieving this OD610 in only 8 hours, indicating a higher development index.
Table 2 shows the percentages of furfural removal by bacterial consortium in 24 hours for different initial concentrations. For lower concentrations, (722 mg L-1, 965 mg L-1, 1365 mg L-1 and 1488 mg L-1) the degradation was 100%. This shows consistency with the increase in bacterial growth (Figure 4), while for 1749 mg L-1, 1991 mg L-1 it was 74%. Finally, for higher concentrations (2724 mg L-1 and 4517 mg L-1) degradation values were only 21% and 11%, respectively.
610 nm absorbance measurements at the initial time and day 1 at different furfural concentrations, incubation conditions: 30 º C at 200 rpm; c0 indicates initial furfural concentratins
. Furfural concentrations on days 0 and 1 and removal percentage for the 8 tests
|
Test |
Concentration (ppm) |
% Furfural Removal |
|
|
day 0 |
day 1 |
|
|
|
1 |
722 |
0 |
100 |
|
2 |
965 |
0 |
100 |
|
3 |
1365 |
0 |
100 |
|
4 |
1488 |
0 |
100 |
|
5 |
1749 |
459 |
74 |
|
6 |
1991 |
512 |
74 |
|
7 |
2724 |
2147 |
21 |
|
8 |
4517 |
4030 |
11 |
Boopathy et al. [38] worked with strains of enteric bacteria. They found similar results, both in biodegradation percentages and bacterial growth, at 960 mg L-1 of furfural. Hunter and Manter [36] showed that the furfural treatments slowed the growth rates of three L. mesenteroides and L. pseudomesenteroidesstrains, but they obtained better degradation values (between 55% to 75% at 4500 mg L-1 of furfural).
It was shown that a bacterial consortium formed by the strains of Microbacterium sp., Brevundimonas sp. and Bacillus licheniformis can use furfural as a carbon source more effectively than the individual strains. In only 48 hours bacterial consortium removes 100% of furfural in concentrations up to 2723 mg L-1. Thus, it could be suggested as an adequate method to eliminate such contaminants.
Furfural concentration influences consortium growth, and prevents the exponential growth of microorganisms during the first 24 hours from 2000 mg L-1.
At high concentrations, such as 4500 mg L-1 of the contaminant, it was verified that there was no exponential bacterial growth within 72 hours and that the percentage of contaminant removal proved to be low.
Future work could focus on immobilizing bacteria techniques and bioreactor assays to improve furfural biodegradation.
|
Abbreviations |
|
|
PAHs PCR rRNA M9 OD610 CFU HPLC |
Polycyclic Aromatic Hydrocarbons Polymerase Chain Reaction Ribosomal Nucleic Acid Minimal medium Optical Density at 610 nm Colony Forming Units High Performance Liquid Chromatography |
|
CODCr CBR |
Chemical Oxygen Demand determined by the Dichromate Method Cyclic Biological Reactor |
- Madloom, A., Jabbar, S., and Kadhim, N., Furfural production based cellulosic garbage. Plant Archives, Vol. 19 No. 2, pp 345-350, 2019.
- Zheng, D., Bao, J., Lu, J. and Gao, C., Isolation and characterization of a furfural degrading bacterium Bacillus cereus sp. strain DS1, Current microbiology, Vol. 70, No. 2, pp 199-205, 2015, https://doi.org/10.1007/s00284-014-0702-9
- Igeño, M., Macias, D. and Blanco, R., A case of adaptive laboratory evolution (ALE): biodegradation of furfural by Pseudomonas pseudoalcaligenes CECT 5344, Genes, Vol. 10, No. 7, pp. 499, 2019, https://doi.org/10.3390/genes10070499
- Ran, H., Zhang, J., Gao, Q., Lin, Z. and Bao, J., Analysis of biodegradation performance of furfural and 5- hydroxymethylfurfural by Amorphothecaresinae ZN1, Biotechnology for Biofuels, Vol. 7, No. 1, pp 1–12, 2014, https://doi.org/10.1186/1754-6834-7-51
- Boopathy, R., Anaerobic biotransformation of furfural to furfuryl alcohol by a methanogenic archaebacterium, International Biodeterioration and Biodegradation, Vol. 63, No. 8, pp 1070–1072, 2009, https://doi.org/10.1016/j.ibiod.2009.09.011
- Crigler, J., Eiteman, M. A. and Altman, E., Characterization of the Furfural and 5-Hydroxymethylfurfural (HMF) Metabolic Pathway in the Novel Isolate Pseudomonas putida ALS1267, Applied Biochemistry and Biotechnology, Vol. 190, No. 3, pp 918-930, 2020, https://doi.org/10.1007/s12010-019-03130-x
- Fernández‐Cabezón, L., Cros, A. and Nikel, P. I., Evolutionary approaches for engineering industrially relevant phenotypes in bacterial cell factories, Biotechnology journal, Vol. 14, No. 9, pp 1800439, 2019, http://dx.doi.org/10.1002/biot.201800439
- Hernández, A.S., Biodiversity of actinomycetes isolated from sewage treatment plants. Study of the degradation capacity of toxic compounds (in Spanish), Ph.D Thesis, Universidad Politécnica de Valencia, España, 2012.
- Benimeli, C., Biodegradation of Organochlorine Pesticides by Aquatic Actinomycetes, Ph.D Thesis, Tucumán National University, Tucumán, Argentina, 2004.
- Polti, M., Study of Chromium Resistance Mechanisms by Regional Actinomycetes Strains, Ph.D Thesis, Tucumán National University, Tucumán, Argentina, 2008.
- Khan M., Ashar N., Ganesh A., Rais N., Faheem S. and Khan S., Bacterial community structure in anaerobic digesters of a full-scale municipal wastewater treatment plant ‒ case study of Dubai, United Arab Emirates, Journal of Sustainable Development of Energy, Water and Environment Systems, Vol. 7, No. 3, pp 385-398, 2019, http://dx.doi.org/10.13044/j.sdewes.d6.0222
- Haleyur N., Shahsavari E., Jain S., Koshlaf E., Ravindran V., Morrison P., Osborn A. and Ball A., Influence of bioaugmentation and biostimulation on PAH degradation in aged contaminated soils: Response and dynamics of the bacterial community, Journal of Environmental Management, Vol. 238, pp 49-58, 2019, https://doi.org/10.1016/j.jenvman.2019.02.115
- Petersohn A., Brigulla M., Haas S., Hoheisel J., Völker U. and Hecker M., Global Analysis of the General Stress Response of Bacillus subtilis, Journal of Bacteriology, Vol. 183, pp 5617–5631, 2001, https://doi.org/10.1128/JB.183.19.5617-5631.2001
- Aspholm, M. E. et al, Biochemical and mutational analysis of spore cortex-lytic enzymes in the food spoiler Bacillus licheniformis, Food Microbiology, Vol. 84, 2019, https://doi.org/10.1016/j.fm.2019.103259
- Reyes-Reyes, M. A., Puentes-Cala E. A., Casanova-Montes E. L., Lopez-Deluque F, Panqueva-Alvarez J.H. and Castillo-Villamizar G. A., Immobilization of potentially crude oil degrading bacteria in natural and synthetic organic matrices, Revista Internacional de Contaminación Ambiental, Vol. 34, No. 4, pp. 597-609, 2018, https://doi.org/10.20937/RICA.2018.34.04.04
- Chirre Flores, J., Patiño, G. A. and Erazo, E. R., Study of the biodegradation of lubricating oil residues retained in bentonite using the bacterial consortium oil eating microbes (Rhodococcus, Pseudomonas and Bacillus), Journal of the Peruvian Chemical Society, Vol. 85, No. 2. pp 163-174, 2019, https://doi.org/10.37761/rsqp.v85i2.75
- Sari G., Trihadiningrum Y. and Ni’matuzahroh, Bioremediation of Petroleum Hydrocarbons in Crude Oil Contaminated Soil from Wonocolo Public Oilfields using Aerobic Composting with Yard Waste and Rumen Residue Amendments, Journal of Sustainable Development of Energy, Water and Environment Systems, Vol. 7, No. 3, pp 482-492, 2019, http://dx.doi.org/10.13044/j.sdewes.d7.0262
- Felshia, S. C., Karthick N. A., Thilagam R., Chandralekha A., Raghavarao K.S.M.S. and Gnanamani A., Efficacy of free and encapsulated Bacillus licheniformis strain SL10 on degradation of phenol: A comparative study of degradation kinetics, Journal of Environmental Management, Vol. 197, pp 373-383, 2017, https://doi.org/10.1016/j.jenvman.2017.04.005
- Yan, Y., Bu, C., He, Q., Zhenga, Z. and Ouyang, J., Efficient bioconversion of furfural to furfuryl alcohol by Bacillus coagulans NL01, Royal Society of Chemistry Advances, Vol. 47, No. 8, pp 26720-26727, 2018, https://doi.org/10.1039/c8ra05098h
- Macaulay, B.M. and Rees, D., Bioremediation of oil spills: a review of challenges for research advancement, Annals of Environmental Science, Vol. 8, pp 9-37, 2014.
- Wang, Z., Yang, Y., Sun, W., Xie, S. and Liu, Y., Nonylphenol biodegradation in river sediment and associated shifts in community structures of bacteria and ammonia-oxidizing microorganisms, Ecotoxicology and Environmental Safety, Vol. 106, pp 1-5, 2014, https://doi.org/10.1016/j.ecoenv.2014.04.019
- Wang X., Wang X., Liu M., Zhou L., Gu Z. and Zhao J., Bioremediation of marine oil pollution by Brevundimonas diminuta: effect of salinity and nutrients, Desalination and Water Treatment, Vol. 57, No. 42, pp 19768-19775, 2016, https://doi.org/10.1080/19443994.2015.1106984
- Viñas, M., Sabaté, J., Espuny, M.J. and Solanas, A.M., Bacterial Community Dynamics and Polycyclic Aromatic Hydrocarbon Degradation during Bioremediation of Heavily Creosote-Contaminated Soil, Applied and Environmental Microbiology, Vol. 71, No. 11, pp 7008-7018, 2005, https://doi.org/10.1128/AEM.71.11.7008-7018.2005
- Basuki, W., Biodegradation of Used Synthetic Lubricating Oil by Brevundimonas diminuta AKL 1.6, Makara Journal of Science, Vol. 21, No. 3, pp 136-142, 2017, https://doi/org/10.7454/mss.v21i3.7382
- Utgés, E. M., Farías, A., Utgés, E. E., Tenev, M., Hervot, E., Nocenti, M. and Echeverría, M., Isolation of native bacteria for later use in the treatment of effluents with furfural, Proceedings of Water Management and Integral Treatment International Congress, (in Spanish), Córdoba, Argentina, pp 1-12, November 9-11, 2015.
- Farías, A., Utgés, E. E., Tenev, M., Hervot, E., Utgés, E. M. and Nocenti, M., Treatment of an effluent with furfural through native bacteria, in an anaerobic fluidized bed reactor, Journal Tecnología y Ciencia UTN (in Spanish), No. 38, pp 18-36, 2020, https://doi.org/10.33414/rtyc.38.18-36.2020
- Echeverría, M., Fontana, G., Utgés, E. M., Cuadra, P. and Temporale, A., Biodegradation of furfural through the use of Microbacterium sp., Proceedings of VII International Congress on Management and Integral Water Treatment (in Spanish), Córdoba, Argentina, pp 1-9, November 14-16, 2018.
- Farías, A., Echeverría, M., Utgés, E. M. and Fontana, G., Bacillus licheniformis as a furfural biodegradation agent and biofilm former, Proceedings of VII Biannual PROIMCA V Biannual PRODECA Congress, (in Spanish), Rafaela, Argentina, pp 660-665, August 7-9, 2019.
- Zheng, D., Bao, J., Lu, J., and Lv, Q., Biodegradation of furfural by Bacillus subtilis strain DS3, Journal of Environmental Biology, Vol. 36, pp 727-732, 2015.
- Bariani, M., Boix, E., Cassella, F. and Cabrera, M. N, Furfural production from rice husks within a biorefinery framework, Biomass Conversion and Biorefinery, pp 1-14, 2020, https://doi.org/10.1007/s13399-020-00810-1
- Steinbach D., Kruse A. and Sauer J., Pretreatment technologies of lignocellulosic biomass in water in view of furfural and 5- hydroxymethylfurfural production- a review, Biomass Convers Biorefinery, Vol. 7, pp 247–274, 2017, https://doi.org/10.1007/s13399-017-0243-0
- Ding, C. Q., Li, K. R., Duan, Y. X., Jia, S. R., Lv, H. X., Bai, H. and Zhong, C., Study on community structure of microbial consortium for the degradation of viscose fiber wastewater, Bioresources and Bioprocessing, 2017, https://doi.org/10.1186/s40643-017-0159-3
- Jing, W., Guiwen, Y., Mingquan, A., Jieli, L., Houming, Z. and Yun, C., Study of a plugging microbial consortium using crude oil as sole carbon source, Petroleum Science, Vol. 5, No. 4 pp 367–374, 2008, https://doi.org/10.1007/s12182-008-0061-x
- Jing, W., Hongke, X. and Shaohui, G., Isolation and Characteristics of a Microbial Consortium for Effectively Degrading Phenanthrene, Petroleum Science Vol. 4, No. 3, 2007, https://doi.org/10.1007/s12182-007-0012-y
- Moussavi, G., Leili, M. and Nadafi, K., Investigation of furfural biodegradation in a continuous inflow cyclic biological reactor, Water Science and Technology, Vol. 73, No. 2, pp 292-301, 2016, https://doi.org/10.2166/wst.2015.488
- Hunter, W. and Manter, D., Pre-treatment step with Leuconostoc mesenteroides or L. pseudomesenteroides strains removes furfural from Zymomonas mobilis ethanolic fermentation broth, Bioresource Technology, Vol. 169, pp 162–168, 2014, https://doi.org/10.1016/j.biortech.2014.06.097
- Guarnieri, M., Franden, M., Johnson, C. and Beckham, G., Conversion and assimilation of furfural and 5-(hydroxymethyl) furfural by Pseudomonas putida KT2440, Metabolic Engineering Communications, Vol. 4, pp 22–28, 2017, https://doi.org/10.1016/j.meteno.2017.02.001
- Boopathy, R., Bokang, H. and Daniels, L., Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria, Journal of Industrial Microbiology, Vol. 11, No. 3, pp 147-150, 1993, https://doi.org/10.1007/BF01583715




